Project 1: Analysis of Cell Autonomous Mechanisms of Phenotypic Plasticity in Invasive Cell Spread
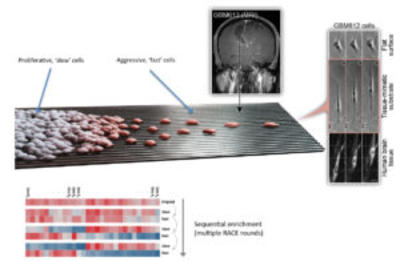
The goal of this project is to obtain a better quantitative understanding of the complex regulation and characteristic of the invasive phenotypic state. The analysis in the project is aimed at the cell autonomous view of regulation, without explicit emphasis of cell-cell interactions. We will combine a variety of tools and approaches, including combining experimentation and mathematical modeling.
We are able to separate slow moving, proliferative cells from fast moving, migratory cells (and enrich each phenotype) using a “phenotypic filter” through an assay called Rapid Analysis of Cell migration Enhancement (RACE). This assay employs nano-fabricated surfaces that result in rapid and highly oriented cell migration. Preliminary results using RACE not only suggest that the assay can help identify molecular mechanisms of the Proliferative-to-Aggressive phenotypic switch but have also led us to develop a preliminary mathematical model describing how specific kinases, thought to be activated by the same growth factors, can be differentially active in different cell sub-populations. In addition, using synthetic biology techniques, we have created a new platform for generating novel kinase inhibitors that can be used to, for the first time, target kinases considered to be “difficult” to target due to their orphan status and unclear regulation mechanisms. When possible, the mechanistic significance in cancer invasion and spreading of newly identified and targeted kinases will be examined using biophysical approaches that can determine whether and how cytoskeletally-mediated processes leading to cell migration have been triggered.
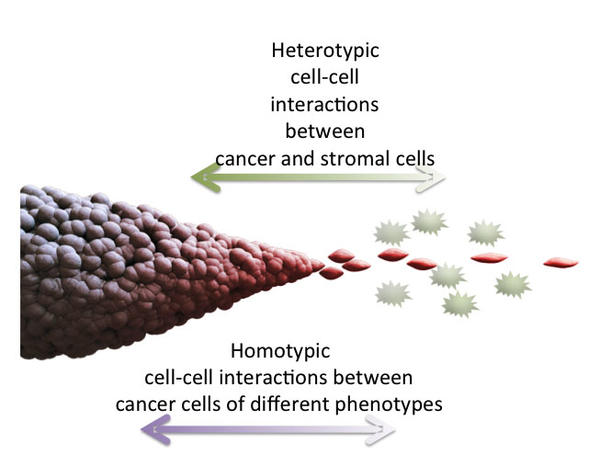 We will examine, in a species-dependent fashion, whether stromal cells exert differential resistance to or cooperation with the invasive cancer spread. A focus will be on the differential expression of specific chemo-attractants and chemo-repellents that are frequently associated with angigogenesis or neuronal migration. We will study the mechanisms regulating communication and putative interaction between cells adopting a more proliferative phenotype with cells adopting a more migratory phenotype. We will use novel techniques and approaches in Project 2 to perturb and validate possible novel targets as regulators of invasive cancer phenotype. Analysis of such targets will be conducted in collaboration with Project 1, since many of the techniques used in these projects are compatible and mutually enriching.
We will examine, in a species-dependent fashion, whether stromal cells exert differential resistance to or cooperation with the invasive cancer spread. A focus will be on the differential expression of specific chemo-attractants and chemo-repellents that are frequently associated with angigogenesis or neuronal migration. We will study the mechanisms regulating communication and putative interaction between cells adopting a more proliferative phenotype with cells adopting a more migratory phenotype. We will use novel techniques and approaches in Project 2 to perturb and validate possible novel targets as regulators of invasive cancer phenotype. Analysis of such targets will be conducted in collaboration with Project 1, since many of the techniques used in these projects are compatible and mutually enriching.